Yoshikazu NAKAMURA, Ph.D.
Institute of Medical Science, University of Tokyo, Minato-ku, Tokyo 108-8639, Japan |
|
Dr. Nakamura is currently a Professor of the Institute of Medical Science, University of Tokyo (IMSUT). Dr. Nakamura received his Ph.D from the University of Kyoto in 1977. He did research as a recipient of a JSPS postdoctoral fellowship at the Institute of Virus Research, Kyoto University (1977-1978) and a postdoctoral research fellowship at the Department of Biology, the University of Utah, USA (1980-1981). He became NCI expert at the National Institutes of Health (NIH), USA, in 1984 and PRI consultant of the Frederick Cancer Research Facility of NIH (1985,1986, 1987, 1988). In 1984 and 1988, he was a visiting scientist at the Institut de Biologie Physico-Chimique in Paris. He was appointed Associated Professor of IMSUT in 1986, then Professor (2000-present), Vice Dean (2002-2003) and Chairman (2003-2007).
Dr. Nakamura is currently Vice President of the Asian-Pacific IMBN (2003- ) and President of the RNA Society of Japan (2006- ), editor of Biochimie (Paris, 1995- ), Associate Editor of Genes to Cells (Oxford, 1998- ), and Editor of Prion (Austin, 2007- ). He was Director of the MEXT project "RNA Network" (2001-2006). He was awarded the Young Investigator Award of the Japanese Biochemical Society (1986) and the Bouboulina Prize (1998).
|
|
|
|

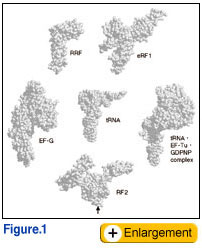
ecent crystal structure and cryo-electron microscopy studies have revealed that proteins called translation factors mimic the shape of tRNA (Fig. 1). One of them, a polypeptide release factor that is required for protein termination, encodes a tripeptide that serves as an ‘anticodon’ to decipher stop codons in mRNA (Ito and Nakamura, 2000; Nakamura et al., 2000). It has long been a puzzle for four decades how protein synthesis terminates at stop codons. These findings undoubtedly solve this long-standing coding problem in the genetic code, and greatly contribute to establishing a novel concept of macromolecular mimicry between protein and RNA. Nature must have evolved this 'art' of molecular mimicry between protein and ribonucleic acid using different protein architectures that are functionally active in a ribosome ‘machine’ (Nakamura and Ito, 2003).
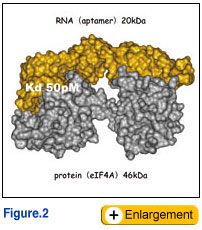
 We aimed to prove this concept by creating novel RNA molecules that could mimic proteins of interest. The systematic evolution of ligands by exponential enrichment (SELEX) method is based on in vitro selection of oligo-nucleotide ligands from large random-sequence libraries by repeated reactions of DNA transcription, RNA selection and RT-PCR amplification (Tuerk and Gold, 1990). The selected oligonucleotide ligands, having both high affinity and specificity to target molecules, are called ‘aptamers’. We have initiated SELEX experiments using mammalian translation initiation factors including eIF4E (Mochizuki et al., 2005), eIF4G (Miyakawa et al., 2006), eIF1A (Oguro and Nakamura, unpublished) and eIF4A (Oguro et al., 2003). Selected RNA aptamers against these target proteins acquired several properties equivalent to, or more importantly, superior to antibodies. One of these aptamers had a Kd on the picomolar scale, an affinity which is a thousand-times stronger than normal antibody. Molecular and biochemical analysis revealed that these aptamers need to be >50 nucleotides long for specific and high affinity binding to their target proteins. Therefore, it might be argued that RNA aptamers to proteins without RNA recognition motifs or strong affinity to RNA can achieve specific high affinity to the target protein by capturing its global conformation (Fig. 2). Ongoing structural studies by NMR and X-ray crystallography support this view. This is completely different from the pinpoint (i.e., epitope <10 amino acids) recognition of target protein by antibodies. This provides us with a solid or promising basis to take steps forward to create novel RNA molecules with distinct structures, which should be equivalent or superior to antibody. We aimed to prove this concept by creating novel RNA molecules that could mimic proteins of interest. The systematic evolution of ligands by exponential enrichment (SELEX) method is based on in vitro selection of oligo-nucleotide ligands from large random-sequence libraries by repeated reactions of DNA transcription, RNA selection and RT-PCR amplification (Tuerk and Gold, 1990). The selected oligonucleotide ligands, having both high affinity and specificity to target molecules, are called ‘aptamers’. We have initiated SELEX experiments using mammalian translation initiation factors including eIF4E (Mochizuki et al., 2005), eIF4G (Miyakawa et al., 2006), eIF1A (Oguro and Nakamura, unpublished) and eIF4A (Oguro et al., 2003). Selected RNA aptamers against these target proteins acquired several properties equivalent to, or more importantly, superior to antibodies. One of these aptamers had a Kd on the picomolar scale, an affinity which is a thousand-times stronger than normal antibody. Molecular and biochemical analysis revealed that these aptamers need to be >50 nucleotides long for specific and high affinity binding to their target proteins. Therefore, it might be argued that RNA aptamers to proteins without RNA recognition motifs or strong affinity to RNA can achieve specific high affinity to the target protein by capturing its global conformation (Fig. 2). Ongoing structural studies by NMR and X-ray crystallography support this view. This is completely different from the pinpoint (i.e., epitope <10 amino acids) recognition of target protein by antibodies. This provides us with a solid or promising basis to take steps forward to create novel RNA molecules with distinct structures, which should be equivalent or superior to antibody.
 Aptamers are artificial molecules isolated from large combinatorial nucleic acid libraries by their high affinity to a target molecule. There are however, several unique features of aptamers that are of additional benefit. Aptamers are not potentially immunogenic and mostly not toxic to cells. They are robust against both reducing conditions and heat denaturation. Finally, they are chemically synthesizable and their antidote molecules can be easily designed. Thus, the application of aptamers in molecular biology and medical science has been extensively explored. In fact, such a dream came true. Pegaptanib (Macugen), the first aptamer-based therapy targeting VEGF (vascular endothelial growth factor) to block abnormal blood vessels to grow and leak, was approved by the United States Food and Drug Administration (FDA) as the therapy for the treatment of age-related macular degeneration (AMD) in December 2004. Aptamers are artificial molecules isolated from large combinatorial nucleic acid libraries by their high affinity to a target molecule. There are however, several unique features of aptamers that are of additional benefit. Aptamers are not potentially immunogenic and mostly not toxic to cells. They are robust against both reducing conditions and heat denaturation. Finally, they are chemically synthesizable and their antidote molecules can be easily designed. Thus, the application of aptamers in molecular biology and medical science has been extensively explored. In fact, such a dream came true. Pegaptanib (Macugen), the first aptamer-based therapy targeting VEGF (vascular endothelial growth factor) to block abnormal blood vessels to grow and leak, was approved by the United States Food and Drug Administration (FDA) as the therapy for the treatment of age-related macular degeneration (AMD) in December 2004.
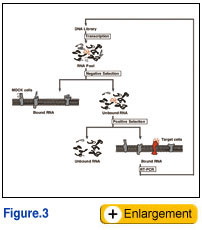 The SELEX is mostly and typically carried out using purified (recombinant) target molecules, and thereby the resulting aptamers are not always reactive to the native targets in the native conditions. This is the big drawback to the current SELEX procedure, especially, for the receptor proteins on cell surface. To overcome this problem, we and others have succeeded the generation of aptamers using live cells as targets. Such aptamers, reactive with native targets on live cells, have large advantages compared with canonical aptamers generated against purified targets. Here, we develop a novel SELEX procedure (referred to as TECS-SELEX) in which cell-surface displayed recombinant protein is directly used as the selection target (Fig. 3). Using this method, we isolated RNA aptamers against transforming growth factor-beta type III receptor expressed on Chinese hamster ovary (CHO) cells (Ohuchi et al. 2006). One of the RNA aptamers has a dissociation constant in the 1 nM range and competed with transforming growth factor-beta to bind to the cell surface receptor in vitro. Thus, TECS-SELEX is a powerful tool that is potentially applicable to any receptors of interest. The SELEX is mostly and typically carried out using purified (recombinant) target molecules, and thereby the resulting aptamers are not always reactive to the native targets in the native conditions. This is the big drawback to the current SELEX procedure, especially, for the receptor proteins on cell surface. To overcome this problem, we and others have succeeded the generation of aptamers using live cells as targets. Such aptamers, reactive with native targets on live cells, have large advantages compared with canonical aptamers generated against purified targets. Here, we develop a novel SELEX procedure (referred to as TECS-SELEX) in which cell-surface displayed recombinant protein is directly used as the selection target (Fig. 3). Using this method, we isolated RNA aptamers against transforming growth factor-beta type III receptor expressed on Chinese hamster ovary (CHO) cells (Ohuchi et al. 2006). One of the RNA aptamers has a dissociation constant in the 1 nM range and competed with transforming growth factor-beta to bind to the cell surface receptor in vitro. Thus, TECS-SELEX is a powerful tool that is potentially applicable to any receptors of interest.
The most advanced RNA aptamer, i.e., close to the clinical study, developed in this laboratory is one raised against midkine (MK), a heparin-binding growth factor involved in oncogenesis, inflammation, and tissue repair. CD4+CD25+ regulatory T (Treg) cells are crucial mediators of autoimmune tolerance. Although the factors that regulate Treg cells are largely unknown, MK-deficient mice are known to resistant to experimental autoimmune encephalomyelitis (EAE). Administration of anti-MK RNA aptamers significantly expanded the Treg cell population and alleviated the symptoms of EAE. These observations indicate that MK serves as a critical suppressor of Treg cells, and inhibition of MK using RNA aptamers may provide an effective therapeutic strategy against autoimmune diseases, including multiple sclerosis (these studies were done in collaboration with Dr. Akio Suzumura, Nagoya University, and Ribomic Inc.).
RNA no longer stands behind DNA or protein but stands in front of them. Recent achievements and discovery in biological science clearly emphasized the importance of RNA in life; the discovery of RNA interference, molecular mimicry between protein and RNA (topics of this lecture), and ribosome structure at atomic resolution. Moreover, the completed human genome project revealed, to our great surprise, the existence of a large amount of protein-noncoding RNAs (ncRNAs). These ncRNAs can be classified into two types: one, like antisense and microRNA, those function with the sequence complementarity to the target mRNA or DNA, while the other, like aptamer, those function independent of the sequence complementarity. We believe that the former class RNA should be a piece of iceberg of ncRNA. Rather, the latter class RNA, e.g., “natural aptamer”, could be major players of ncRNAs. By studying these natural and artificial RNA aptamers, we hope to clarify superior potential of RNA, which would be highly beneficial to the development of RNA medicine and the comprehensive understanding of human genome RNA function.
|
|
 |
|
Reference: |
|
| 1. |
Ito, K., Uno, M., Nakamura, Y.: A tripeptide 'anticodon' deciphers stop codons in messenger RNA. Nature, 403: 680-684 (2000) |
| 2. |
Miyakawa, S., Oguro, A., Ohtsu, T., Imataka, H., Sonenberg, N., Nakamura, Y.: RNA aptamers to mammalian initiation factor 4G inhibit cap-dependent translation by blocking the formation of initiation factor complexes. RNA, 12: 1825-1834 (2006). |
| 3. |
Mochizuki, K., Oguro, A., Ohtsu, T., Sonenberg, N., Nakamura, Y.: High affinity RNA for mammalian initiation factor 4E interferes with mRNA-cap binding and inhibits translation. RNA, 11: 77-89 (2005). |
| 4. |
Nakamura, Y., Ito, K.: Making sense of mimic in translation termination. Trends Biochem. Sci., 28: 99-105 (2003). |
| 5. |
Nakamura, Y., Ito, K., Ehrenberg, M.: Mimicry grasps reality in translation termination. Cell, 101: 349-352 (2000). |
| 6. |
Oguro, A., Ohtsu, T., Svitkin, Y., Sonenberg, N., Nakamura, Y.: RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA, 9: 394-407 (2003). |
| 7. |
Ohuchi, S.P., Ohtsu, T., Nakamura, Y.: Selection of RNA aptamers against recombinant transforming growth factor-beta type III receptor displayed on cell surface. Biochimie, 88: 897-904 (2006). |
| 8. |
Tuerk, C. and Gold, L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DAN polymerase. Science 249: 505-510. |
|
|


